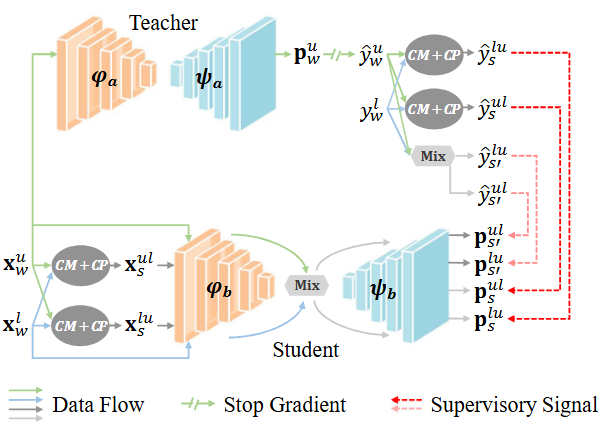
This repository contains the official PyTorch implementation of the paper Cross-Set Data Augmentation for Semi-Supervised Medical Image Segmentation
Medical image semantic segmentation is a fundamental yet challenging research task. However, training a fully supervised model for this task requires a substantial amount of pixel-level annotated data, which poses a significant challenge for annotators due to the necessity of specialized medical expert knowledge. To mitigate the labeling burden, a semi-supervised medical image segmentation model that leverages both a small quantity of labeled data and a substantial amount of unlabeled data has attracted prominent attention. However, the performance of current methods is constrained by the distribution mismatch problem between limited labeled and unlabeled datasets. To address this issue, we propose a cross-set data augmentation strategy aimed at minimizing the feature divergence between labeled and unlabeled data. Our approach involves mixing labeled and unlabeled data, as well as integrating ground truth with pseudo-labels to produce augmented samples. By employing three distinct cross-set data augmentation strategies, we enhance the diversity of the training dataset and fully exploit the perturbation space. Our experimental results on COVID-19 CT data, pinal cord gray matter MRI data and prostate T2-weighted MRI data substantiate the efficacy of our proposed approach.
-
We recommend installing the environment through conda and pip
-
For experiments on COVID-19-20 dataset
cd CDA/exps_on_COVID-19-20/code/ # Create a conda environment (python version >= 3.8) conda create -n covid python=3.8 # Activate the environment conda activate covid # Install torch (torch version >= 1.11.0, cuda version >= 1.13) and torchvision # Please refer to https://pytorch.org/ if you need a different cuda version pip install torch==1.11.0+cu113 torchvision==0.12.0+cu113 torchaudio==0.11.0 --extra-index-url https://download.pytorch.org/whl/cu113 # Install dependencies pip install -r requirements.txt
-
For experiments on SCGM dataset
cd CDA/exps_on_SCGM/code/ # Install dependencies and conda environment conda env create -f semi_dg_cda.yaml # Activate the environment conda activate semidg_cda
-
COVID-19-20
-
We followed the settings of SASSL and the pre-processed dataset can be downloaded from the link.
-
Extract the sets to
$ROOT/exps_on_COVID-19-20/data. The directory structure should look like as follows:$ROOT/exps_on_COVID-19-20/data/ ├── COVID249/ │ ├── NII/ (Original dataset in NIFTI) │ ├── PNG/ (Pre-processed dataset in PNG) │ ├── train_0.1_l.xlsx (Datasplit for 10% setting) │ ├── train_0.1_u.xlsx (Datasplit for 10% setting) │ ├── train_0.2_l.xlsx (Datasplit for 20% setting) │ ├── train_0.2_u.xlsx (Datasplit for 20% setting) │ ├── train_0.3_l.xlsx (Datasplit for 30% setting) │ ├── train_0.3_u.xlsx (Datasplit for 30% setting) │ ├── test_slice.xlsx (Datasplit for testing) │ ├── val_slice.xlsx (Datasplit for validation)
-
-
SCGM
-
We followed the settings of EPL and the original dataset can be downloaded from the official website.
-
Extract the training and testing data to
$ROOT/exps_on_SCGM/data/scgm_rawdata/trainand$ROOT/exps_on_SCGM/data/scgm_rawdata/test, respectively. -
You need first to change the dirs (lines 31 to 52) in the scripts exps_on_SCGM/data/preprocess/save_SCGM_2D.py, and then run
save_SCGM_2D.pyto split the original dataset into labeled and unlabeled sets in four domains. -
The directory structure should look like as follows:
$ROOT/exps_on_SCGM/data/ ├── scgm/ │ ├── scgm_split_2D_data/ (Pre-processed image in NUMPY ARRAYS) │ ├── Labeled/ (Labeled data in four domains) │ ├── vendorA/ (Datasplit for domain UCL) │ ├── 000000.npz │ ├── ... │ ├── vendorB/ (Datasplit for domain Montreal) │ ├── vendorC/ (Datasplit for domain Zurich) │ ├── vendorD/ (Datasplit for domain Vanderbilt) │ ├── Unlabeled/ (Unlabeled data in four domains) │ ├── scgm_split_2D_mask/ (Pre-processed segmentation masks in NUMPY ARRAYS) │ ├── Labeled/ (Labeled masks in four domains)
-
-
For experiments on COVID-19-20
- To train our
CDAwith different labeled ratios, please refer to lines 22 to 35 in the bash script exps_on_COVID-19-20/code/main.sh. - There are 3 steps:
- Step 1: Extract tumor images and masks. The main interface is implemented in
exps_on_COVID-19-20/code/extract_tumors.py. You need to modify the paths (data_root,file_path,save_dir) in the script for different labeled ratios. We suggest putting all data under the same folder ($ROOT/exps_on_COVID-19-20/data/COVID249/); - Step 2: Pre-train merely using labeled data augmented by tumor copy-paste. The main interface is implemented in
exps_on_COVID-19-20/code/train_baseline_tumorcptransV2.py; - Step 3: train CDA initialized by pre-trained paramters in a semi-supervised segmentation setting. The main interface is implemented in
exps_on_COVID-19-20/code/train_CDA.py.
- Step 1: Extract tumor images and masks. The main interface is implemented in
- Below we provide an example for training
CDAwith10%labeled ratio.-
--root_pathand--expdefine the location to save model weights, along with a log file and a tensorboard file. -
--tumor_dirdenotes the location where tumor data generated in Step 1 are put. -
--dataroot_pathdenotes the location where datasets are put. -
--reload_pathdenotes the location where pre-trained parameters learned in Step 2 are saved. -
--excel_file_name_labeldenotes the split for labeled data, and--excel_file_name_unlabeldenotes the split for unlabeled data. -
--modeldefines the type of model's framework. -
--labeled_perdenotes labeled ratio.cd CDA/exps_on_COVID-19-20/code/ # Step1 # data_root='$ROOT/exps_on_COVID-19-20/data/', file_path='$ROOT/exps_on_COVID-19-20/data/COVID249/train_0.1_l.xlsx', save_dir='$ROOT/exps_on_COVID-19-20/data/COVID249/tumor_0.1/' python extract_tumors.py # Step2 CUDA_VISIBLE_DEVICES=0 python train_baseline_tumorcptransV2.py \ --root_path /root/autodl-fs/CDA/exps_on_COVID-19-20/ \ --tumor_dir /root/autodl-fs/CDA/exps_on_COVID-19-20/data/COVID249/tumor_0.1/ \ --dataroot_path $ROOT/exps_on_COVID-19-20/ \ --excel_file_name_label train_0.1_l.xlsx \ --excel_file_name_unlabel train_0.1_u.xlsx \ --exp baseline_tumorcp_transform_blur \ --labeled_per 0.1 # Step3 CUDA_VISIBLE_DEVICES=0 python train_CDA.py \ --root_path /root/autodl-fs/CDA/exps_on_COVID-19-20/ \ --tumor_dir /root/autodl-fs/CDA/exps_on_COVID-19-20/data/COVID249/tumor_0.1/ \ --reload_path /root/autodl-fs/CDA/exps_on_COVID-19-20/exp/COVID249/exp_baseline_tumorcp_transform_blur_0.1_unet/model_best.pth \ --dataroot_path $ROOT/exps_on_COVID-19-20/ \ --excel_file_name_label train_0.1_l.xlsx --excel_file_name_unlabel train_0.1_u.xlsx --exp CDA \ --model unet_imgcut_feaaugmixup \ --labeled_per 0.1
-
- For other models such as
BaselineandBCP, please refer to exps_on_COVID-19-20/code/main.sh.exps_on_COVID-19-20/code/train_baseline.pyandexps_on_COVID-19-20/code/train_BCP.pyare the implementation of corresponding models.
- To train our
-
For experiments on SCGM
-
To train our
CDAusing 20% labeled data in three domains, please refer to the scrips inexps_on_SCGM/code/CDA. -
There are three steps:
- Step 1: Modify the paths of the dataset. You can change the dirs (lines 27 to 42) in scgm_dataloader.py.
- Step 2: Pre-train merely using labeled data augmented by copy-paste. The main interface is implemented in
train_baseline_instancecp_deeplabv3_epldiceloss_savebestema_xxx.sh, and you can change the path to save a log file in this script.- The config information is defined in
config_scgm_deeplabv3_epldiceloss_xxx.py, - The locations to save model weights and a tensorboard file are defined in
train_baseline_instancecp_deeplabv3_epldiceloss_savebestema_xxx.py.
- The config information is defined in
- Step 3: train CDA initialized by pre-trained paramters in a domain-generalized semi-supervised segmentation setting. The main interface is implemented in
train_CDA_cutmixl2u_instancecpl2u_feamixl2u_deeplabv3_epldiceloss_savebestema_loadselftrainparam_xxx.sh, and the config information is defined inconfig_scgm_cutmixinstancefeamix_deeplabv3_epldiceloss_loadselftrainparam_xxx.py.
-
Below we provide an example for training
CDAusing data in domains B, C, D, and inferencing in domain A.# Step 2 sh train_baseline_instancecp_deeplabv3_epldiceloss_savebestema_A.sh # Step 3 sh train_CDA_cutmixl2u_instancecpl2u_feamixl2u_deeplabv3_epldiceloss_savebestema_loadselftrainparam_A.sh
-
For training the
EPL, please refer to the bash scripts inexps_on_SCGM/code/EPL, and the config information is defined inconfig_scgm_loadaugbaseline_1gpu_xxx.py.
-
-
For experiments on COVID-19-20
-
To run the evaluation code, please refer to lines 39 to 41 in the bash script exps_on_COVID-19-20/code/main.sh.
-
Below we provide an example for testing
CDAwith10%labeled ratio.--model_pathdenotes the location where trained CDA are saved.
CUDA_VISIBLE_DEVICES=0 python test.py \ --root_path /root/autodl-fs/CDA/exps_on_COVID-19-20/test/ \ --model_path /root/autodl-fs/CDA/exps_on_COVID-19-20/exp/COVID249/exp_CDA_0.1_unet_imgcut_feaaugmixup/model_best.pth \ --dataroot_path /root/autodl-fs/CDA/exps_on_COVID-19-20/ \ --exp CDA \ --model unet_imgcut_feaaugmixup \ --labeled_per 0.1
-
For other models such as
BaselineandBCP, you can modify--model_path,--exp, and--model, more details please refer to exps_on_COVID-19-20/code/main.sh.
-
-
For experiments on SCGM
-
To run the evaluation code, please update the paths of saved model weights at line 216 in
exps_on_SCGM/code/CDA/inference_scgm_onemodel.py.- if you need to save the predicted masks, please set
saveimgas True.
- if you need to save the predicted masks, please set
-
Next, run the following command:
python inference_scgm_onemodel.py
-
If you need to calculate the average results of four domains, we provide
exps_on_SCGM/code/CDA/inference_scgm_onemodel_alldomain_avg.py. The path of saved model trained in different domains need to be modified. Next, run the following command:python inference_scgm_onemodel_alldomain_avg.py
-
To evaluate
EPL, similarly, update the paths of saved model weights inexps_on_SCGM/code/EPL/inference_scgm.pyorexps_on_SCGM/code/EPL/inference_scgm_alldomain_avg.py. Next, run the following commands:# Evaluate the performance of a model in a target domain python inference_scgm.py # Evaluate the average performance of model in four target domains python inference_scgm_alldomain_avg.py
-
This repository is based on the codes and datasets provided by the projects: SASSL and EPL.
Thanks a lot for their great works.
If you use this code in your research, please kindly cite the following papers:
@article{wu2024cross,
title={Cross-Set Data Augmentation for Semi-Supervised Medical Image Segmentation},
author={Wu, Qianhao and Jiang, Xixi and Zhang, Dong and Feng, Yifei and Tang, Jinhu},
journal={Image and Vision Computing},
year={2024}
}