-
Notifications
You must be signed in to change notification settings - Fork 57
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Pop reduction #539
Pop reduction #539
Conversation
…pdate`` and ``CustomUpdateWU`` and split into ``isBatchReduction`` and ``isNeuronReduction``
* added warning about bug
…f variable with a delay buffer
Codecov ReportBase: 87.04% // Head: 89.38% // Increases project coverage by
Additional details and impacted files@@ Coverage Diff @@
## master #539 +/- ##
==========================================
+ Coverage 87.04% 89.38% +2.33%
==========================================
Files 84 73 -11
Lines 18099 10747 -7352
==========================================
- Hits 15754 9606 -6148
+ Misses 2345 1141 -1204
Help us with your feedback. Take ten seconds to tell us how you rate us. Have a feature suggestion? Share it here. ☔ View full report at Codecov. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Looking good. Thanks for also updating the documentation.
Since #447, GeNN has supported 'batch reductions' where you can reduce (calculate the sum or maximum) a per-neuron/synapse state variable across the instances of a batched model. This PR introduces a basic implementation of 'neuron reductions' allowing the same reduction operations to be applied across a population of neurons e.g. to implement a softmax function on a population of output neurons. As is all too often the case, the majority of changes relate to refactoring the code that figures out how to index into variables to handle the new cases this PR enables!
Variable duplication modes
Variables that are shared across batched model instances are marked with an access mode containing the
VarAccessDuplication::SHAREDflag . As well as allowing for memory savings, for example by sharing weights, this allows these variables to be used as the targets for batch reductions. Similarly, this PR introduces a newVarAccessDuplication::SHARED_NEURONflag for variables which are shared between all the neurons in a population (but duplicated across batches). Outside of custom updates, the only access mode where this is allowable isVarAccess::READ_ONLY_SHARED_NEURONwhich results in a read-only variable shared between all neurons in a population.Note: this functionality ends up rather duplicating the functionality of non-pointer extra global parameters but this is something I intend to unify in GeNN 5.0.0
Variable access modes
Now, with all the indexing fun required to deal with the new duplication modes, custom updates can now reduce into
VarAccess::READ_ONLY_SHARED_NEURONvariables using theVarAccessMode::REDUCE_SUMorVarAccessMode::REDUCE_MAXaccess modes or define their own state variables with theVarAccess::REDUCE_NEURON_SUMandVarAccess::REDUCE_NEURON_MAXaccess modes.SIMT implementation
Similarly to our batch reductions, I copied the TF algorithm used in the common case (used when number of neurons < 1024). This uses a warp of threads per batch:
This is tailored to e.g. small populations of output neurons (around warp size) and large batch size which can reasonably occupy GPU using this approach. Implementation at https://github.com/genn-team/genn/blob/pop_reduction/src/genn/genn/code_generator/backendSIMT.cc#L934-L988.
Only fly in the ointment is that OpenCL 1.2 does not expose warp-level operations so I have just added an error and left some rather CUDA-specific code generation in the SIMT backend. I can tidy this up another time.
CPU implementation
This is super-simple it just applies the operation in a loop! https://github.com/genn-team/genn/blob/pop_reduction/src/genn/backends/single_threaded_cpu/backend.cc#L562-L586
Syntax
Aside from the flags, no new syntax is required so a sum of the membrane voltages of a population of Izhikevich neurons can be calculated like this:
C++
Python
Finally, see the custom_update_neuron_reduction_batch_one feature test for a more complex where I implement softmax
Performance
To evaluate performance, I replaced the CUDA intrinsics previously used by mlGeNN to implement softmax with a version implemented using this system and trained a single epoch of latency MNIST using eProp. As you might expect, adding three extra kernel launches per-timestep does reduce performance (mostly through CPU overheads):
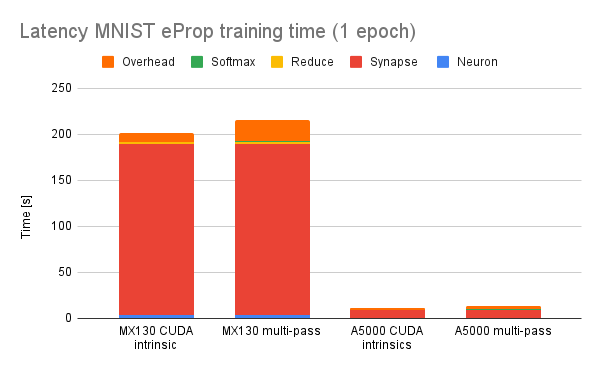
However, this is pretty much the worst case as this is a tiny model with 100 hidden neurons so the (approximately constant) time taken to calculate the reduction is more significant than it would be if e.g. we were using a larger model/more computationally expensive ALIF model.
Future
Fixes #371