├── Readme.md // help
├── alg // method code
│ ├── model.py
│ └── main.py
├── baselines // baseline methods code
│ ├── LSTM
│ ├── SR
│ ├── ECEC
│ ├── EWC
│ ├── GEM
│ ├── ORGFW
│ ├── CLEAR
│ └── CLOPS // https://github.com/danikiyasseh/CLOPS
├── DataProcess // data processing code
│ ├── DataProcess.ipynb
│ ├── DataStatistics.ipynb
│ └── ResultsVis.ipynb
├── data // raw data
├── BatchData // preprocessed data
├── StageData // preprocessed data
├── StageBatchData // preprocessed data for test
├── figures // result figures
├── model // saved DL model
├── results // result files
└── example.ipynb // runnable test case
The RU code requires the following:
- Python 3.6 or higher
- TensorFlow 1.13 or higher
The datasets can be downloaded from the following links:
In order to pre-process the datasets appropriately for RU, please run DataProcess.ipynb and get data in BatchData
To test the model(s) in the paper, run example.ipynb
To train the model(s) in the paper, run this command:
python alg/main.py
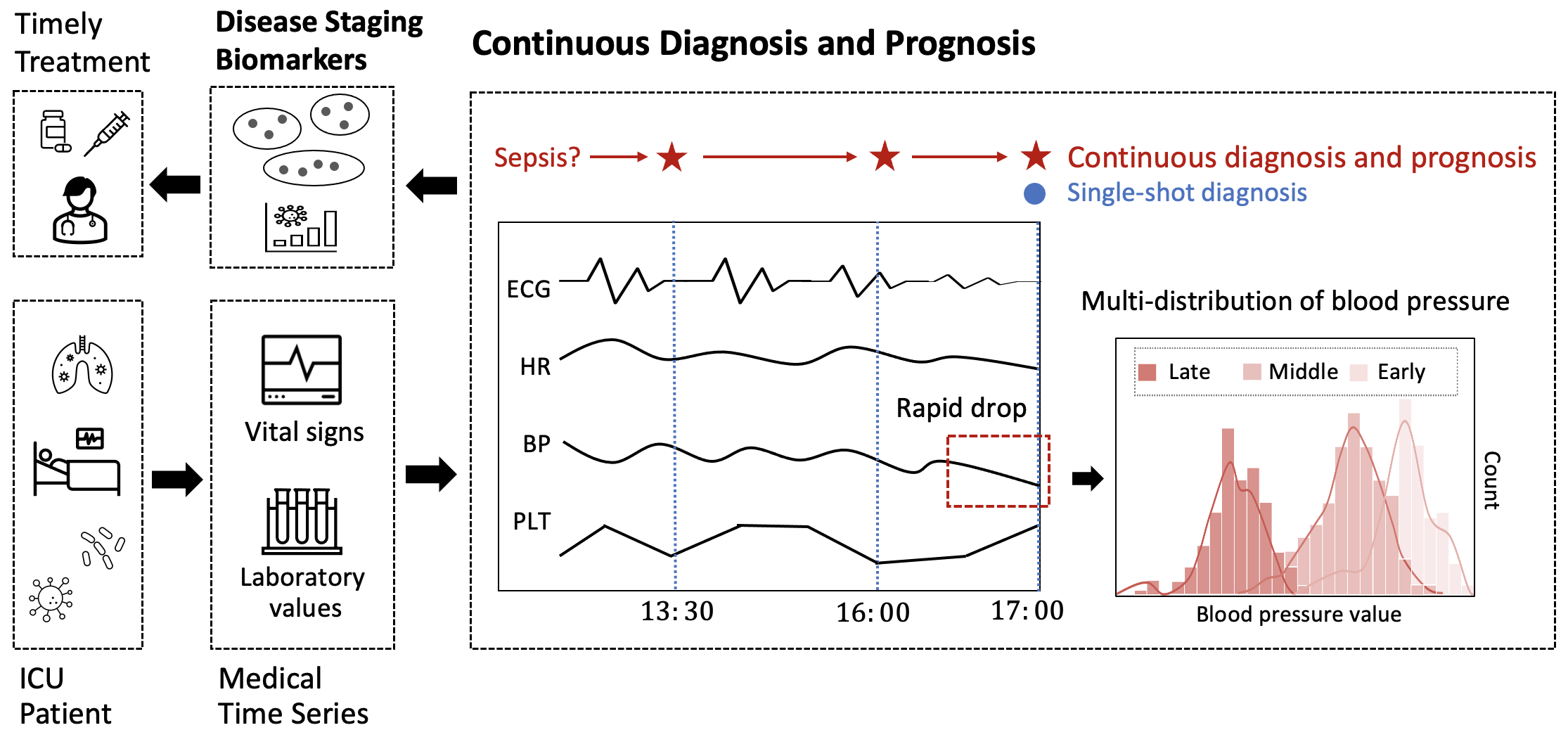
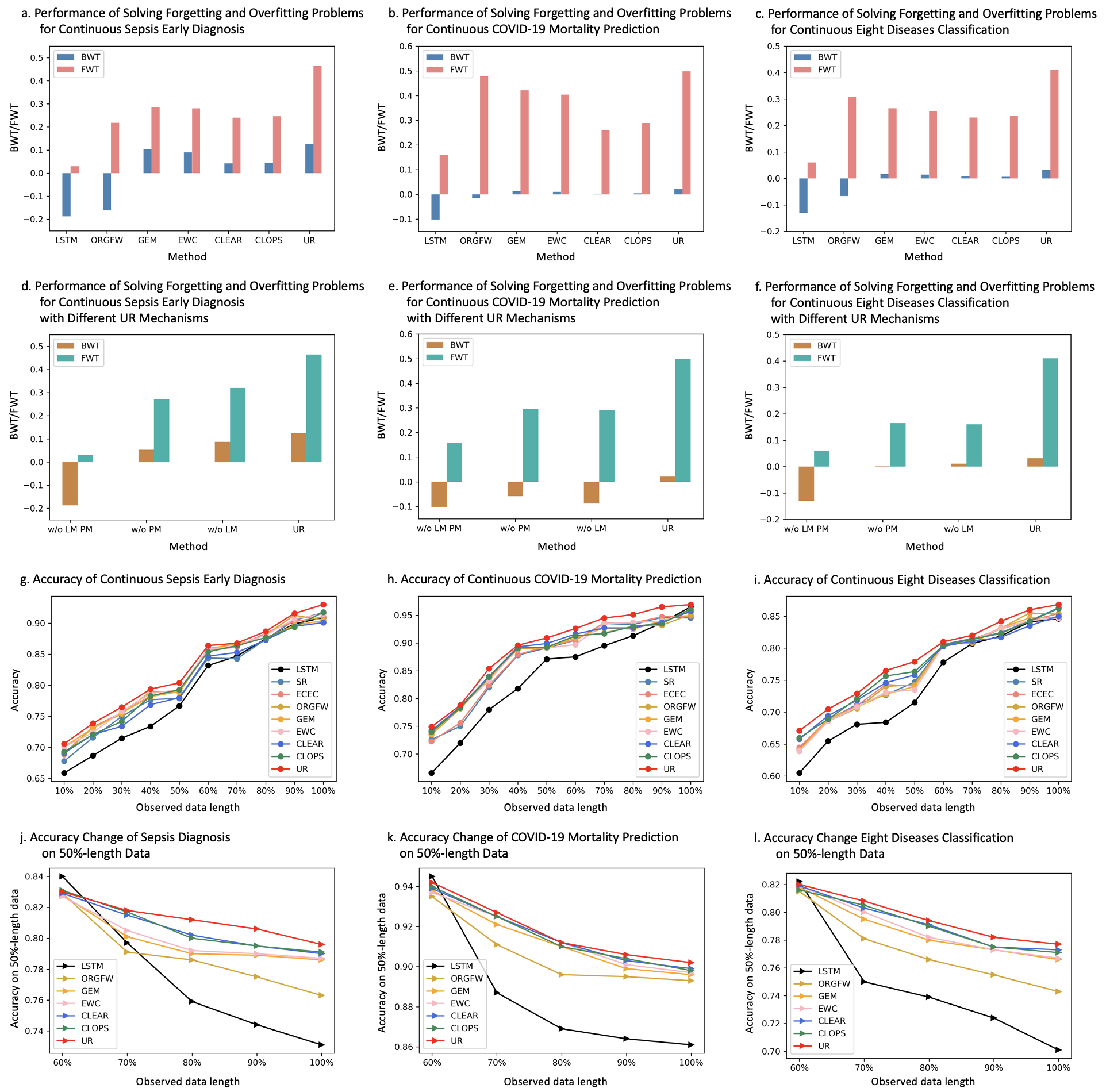
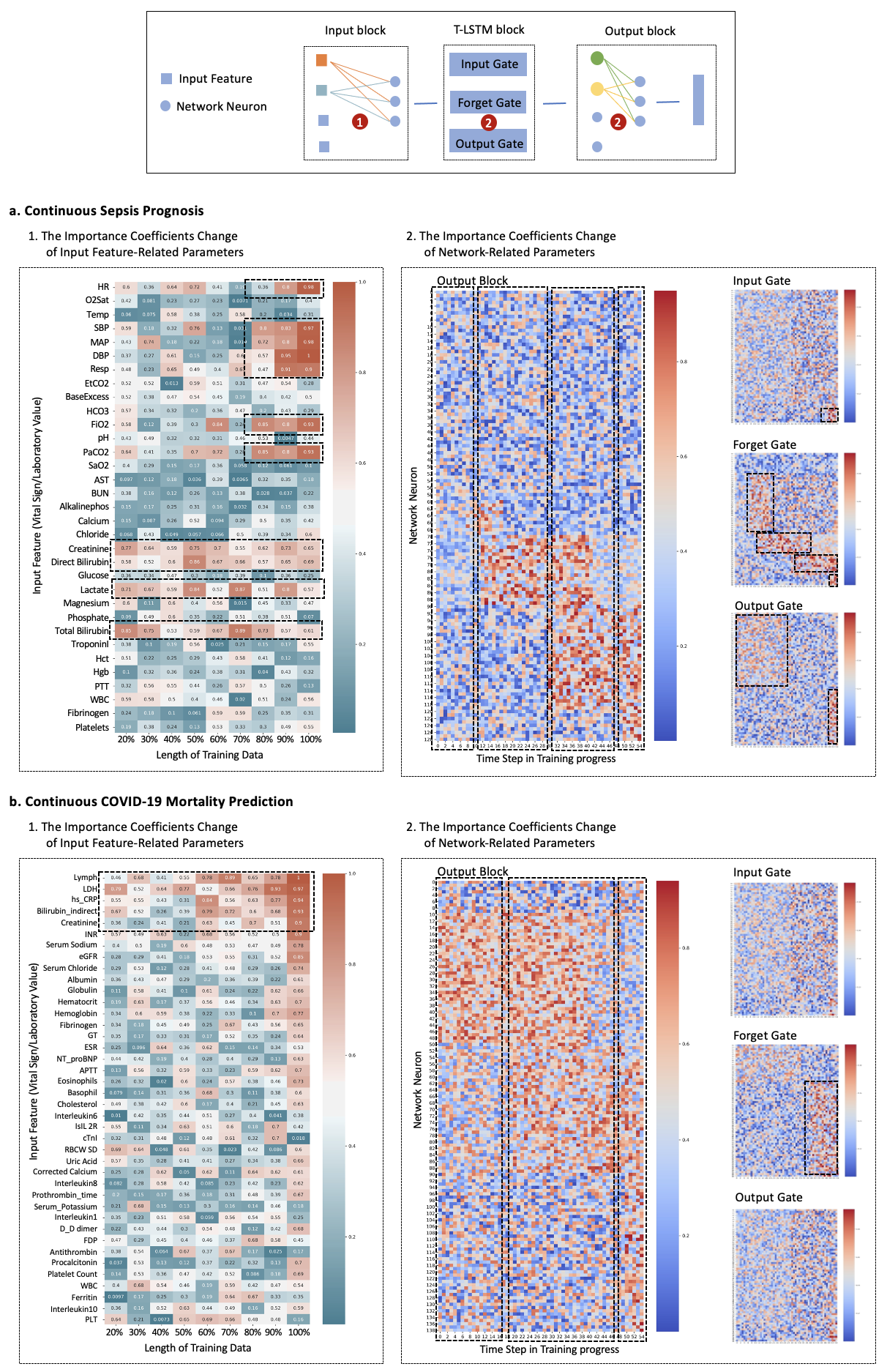
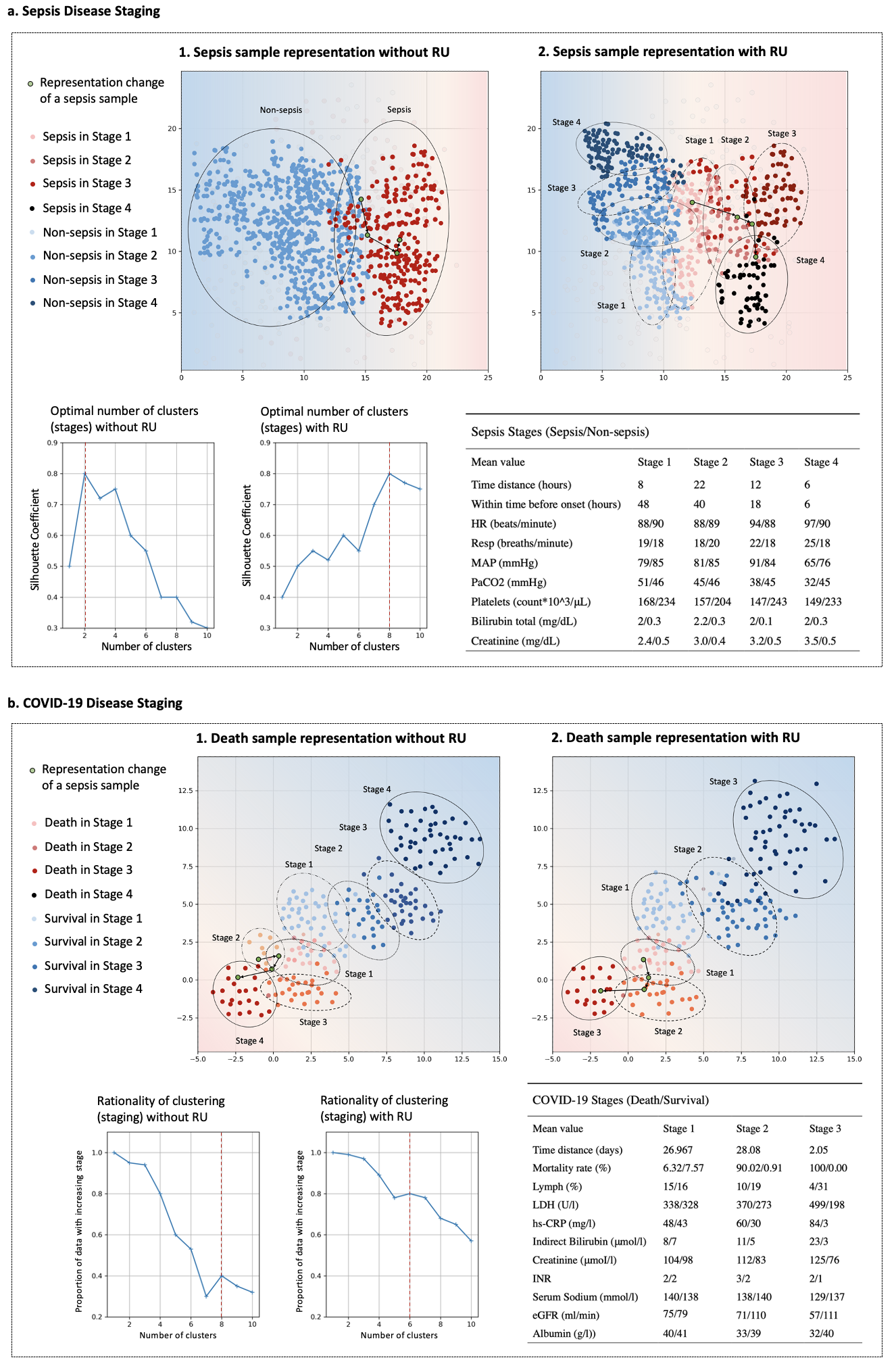
Continuous diagnosis and prognosis are essential for intensive care patients. It can provide more opportunities for timely treatment and rational resource allocation, especially for sepsis, a main cause of death in ICU, and COVID-19, a new worldwide epidemic. Although deep learning (DL) methods have shown their great superiority in many medical tasks, they tend to catastrophically forget, over fit, and get results too late when performing diagnosis and prognosis in the continuous mode. In this manuscript, we achieved high accuracy results for continuous diagnosis and prognosis, 90%, 97%, and 85% accuracy on continuous sepsis prognosis, continuous COVID-19 mortality prediction, and continuous eight diseases classification. We found 4 stages for sepsis with the 6 typical biomarkers (heart rate, respiration mean arterial pressure, PaCO2, platelets count, total bilirubin, and creatinine). We found three stages for COVID-19 with the 5 typical biomarkers (lymphocytes, lactic dehydrogenase, high-sensitivity C-reactive protein, indirect bilirubin, and creatinine).
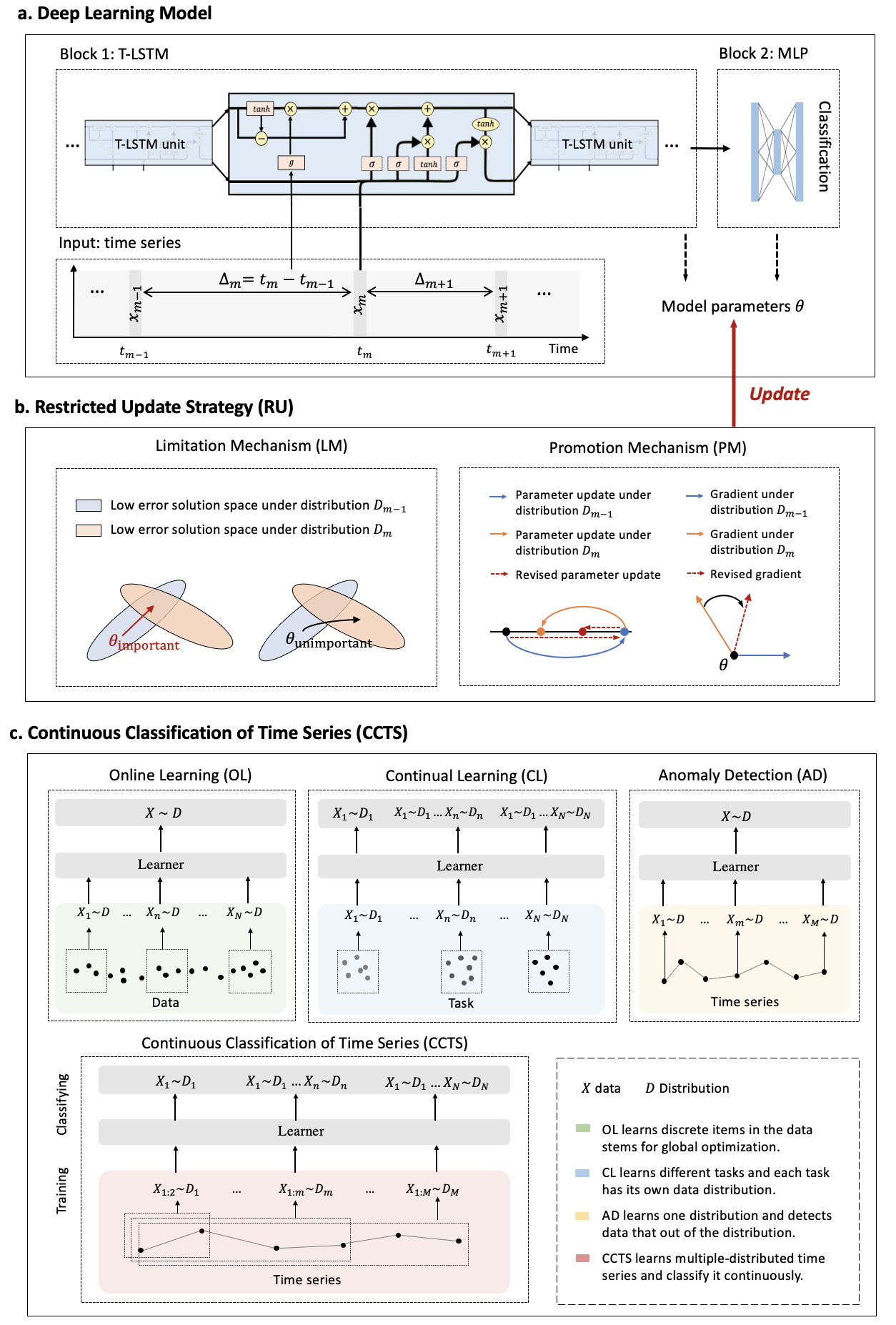
The major advantages of our study are fourfold: (1) For continuous diagnosis and prognosis of time-sensitive illness, we design a restricted update strategy of neural networks (RU) for the DL model, which outperforms baselines. (2) RU has a certain ability to interpret the update of DL models and the change of medical time series through input indicators and parameter visualization. These side effects make our method attractive in medical applications where model interpretation and marker discovery are required. (3) We extend our method to connect the distribution change of vital signs with the parameter change of the DL model and we find typical disease biomarkers and stages of sepsis and COVID-19. (4) RU is a data-agnostic, model-agnostic, and easy-to-use plug-in. It can be used to train various types of DL models. Note that such a continuous prediction mode is needed in most time-sensitive applications, not just in medical tasks. We define these tasks with a new concept, Continues Classification of Time Series (CCTS).