This project to show the genetic algorithm process in python
Why python? cos I love it
hate python? its your problem :P
pip install numpy
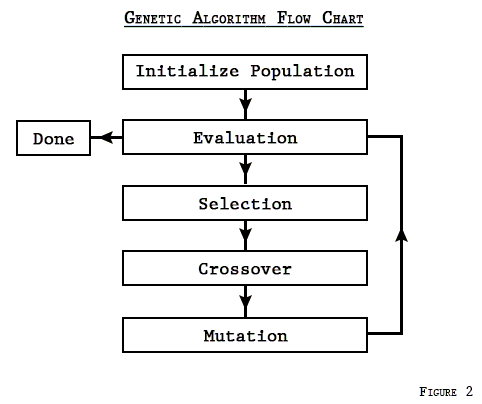
Flowchart of GA
What is population? Population is a collection of genes
What is gen? its an individual in the population
for simple explanation, I have an example :
gen = 'Hello World!'
Population = ('Hello World!', 'Hello Wordd!', 'Hello Morth!')
so how to create or initialize population? just create random gen and collect it save it in a variable. This is the code how to generate new random gen and collect it into a population. The datatype of population is a dict, for matlab just use struct. Before save it into a population we need to calculate the fitness between a gen with the target. If a gen is match with the target, the value of fitness is 100. So in the population we will a pair of gen and its fitness
# generate new gen
def create_gen(panjang_target):
random_number = np.random.randint(32, 126, size=panjang_target)
gen = ''.join([chr(i) for i in random_number])
return gen
# calculate fitness of gen
def calculate_fitness(gen, target, panjang_target):
fitness = 0
for i in range (panjang_target):
if gen[i:i+1] == target[i:i+1]:
fitness += 1
fitness = fitness / panjang_target * 100
return fitness
# create population
def create_population(target, max_population, panjang_target):
populasi = {}
for i in range(max_population):
gen = create_gen(panjang_target)
genfitness = calculate_fitness(gen, target, panjang_target)
populasi[gen] = genfitness
return populasi
Selection is a process to choose 2 best from a population
How to choose it? just check the fitness of each gen and choose 2 biggest in a population
# selection process
def selection(populasi):
pop = dict(populasi)
parent = {}
for i in range(2):
gen = max(pop, key=pop.get)
genfitness = pop[gen]
parent[gen] = genfitness
if i == 0:
del pop[gen]
return parent
crossover is doing sexual activities between 2 parent which choosen in selection process
In this code, crossover process is making process like this :
parent1 = 'abcde12345'
parent2 = '12345abcde'
offspring1 = 'abcdeabcde'
offspring2 = '1234512345'
the idea is getting last half total character from parent1 and combine with first half total character from parent2 and vice versa. The full code of this process is like this
# crossover
def crossover(parent, target, panjang_target):
child = {}
cp = round(len(list(parent)[0])/2)
for i in range(2):
gen = list(parent)[i][:cp] + list(parent)[1-i][cp:]
genfitness = calculate_fitness(gen, target, panjang_target)
child[gen] = genfitness
return child
mutation process is genetic operator used to maintain genetic diversity from one generation of a population of genetic algorithm chromosomes to the next
the idea is we need to choose mutation rate, and looping for every character in a gen to get random number. If the random number is bigger than mutation rate, in that looping character will be replaced with random character
# mutation
def mutation(child, target, mutation_rate, panjang_target):
mutant = {}
for i in range(len(child)):
data = list(list(child)[i])
for j in range(len(data)):
if np.random.rand(1) <= mutation_rate:
ch = chr(np.random.randint(32, 126))
data[j] = ch
gen = ''.join(data)
genfitness = calculate_fitness(gen, target, panjang_target)
mutant[gen] = genfitness
return mutant
evaluation process is check what fitness value
if the fitness of mutation process is equal to 100% so the guess is same with target word and the process will be stoped
if bestfitness(mutant) >= 100:
break
regeneration is insert gen of mutation process into a population
in this process, the worst gen in a population will be dropped and replace with new gen from mutation process
# create new population with new best gen
def regeneration(mutant, populasi):
for i in range(len(mutant)):
bad_gen = min(populasi, key=populasi.get)
del populasi[bad_gen]
populasi.update(mutant)
return populasi