Percentage of association entites that were sucessfully harmonized.
Percentage of association entites that were sucessfully harmonized.
harmonizations = Harmonizations(es_host=args.host, index=args.index)
harmonizations.harmonization_percentages_figure(path='./images/harmonization_percentages.png')
 Percentage of others association genes appear in source.
Percentage of others association genes appear in source.
overlaps = Overlaps(es_host=args.host, index=args.index)
overlaps.gene_overlaps_figure(path='./images/gene_overlaps.png')
 Percentage of others association features appear in source.
Percentage of others association features appear in source.
overlaps = Overlaps(es_host=args.host, index=args.index)
overlaps.feature_overlaps_figure(path='./images/feature_overlaps.png')
 Percentage of others association evidence publications appear in source.
Percentage of others association evidence publications appear in source.
overlaps = Overlaps(es_host=args.host, index=args.index)
overlaps.publication_overlaps_figure(path='./images/publication_overlaps.png')
 Percentage of others association phenotype appear in source.
Percentage of others association phenotype appear in source.
overlaps = Overlaps(es_host=args.host, index=args.index)
overlaps.phenotype_overlaps_figure(path='./images/phenotype_overlaps.png')
 Percentage of others association environment appear in source.
Percentage of others association environment appear in source.
overlaps = Overlaps(es_host=args.host, index=args.index)
overlaps.environment_overlaps_figure(path='./images/environment_overlaps.png')
 Breakdown of top twenty phenotypes in g2p knowledgebases.
Breakdown of top twenty phenotypes in g2p knowledgebases.
cohorts = Cohorts(es_host=args.host, index=args.index)
cohorts.g2p_phenotypes_by_source_figure(path='./images/g2p_phenotypes_by_source.png')
 Show the evidence phenotypes mapped to feature chromosome.
Show the evidence phenotypes mapped to feature chromosome.
cohorts = Cohorts(es_host=args.host, index=args.index)
cohorts.g2p_phenotypes_by_chromosome_figure(path='./images/g2p_phenotypes_by_chromosome.png')
 Show the evidence phenotypes mapped to tumor sample to feature chromosome.
Show the evidence phenotypes mapped to tumor sample to feature chromosome.
cohorts = Cohorts(es_host=args.host, index=args.index)
cohorts.genie_samples_by_chromosome_figure(path='./images/genie_samples_by_chromosome.png')
 Breakdown of top twenty genie clinical phenotypes in g2p knowledgebases.
Breakdown of top twenty genie clinical phenotypes in g2p knowledgebases.
cohorts = Cohorts(es_host=args.host, index=args.index)
cohorts.genie_samples_by_oncotree_figure(path='./images/genie_samples_by_oncotree.png')
 Breakdown of top twenty genie clinical phenotypes by bio marker type.
Breakdown of top twenty genie clinical phenotypes by bio marker type.
cohorts = Cohorts(es_host=args.host, index=args.index)
cohorts.genie_biomarkers_figure(path='./images/genie_biomarkers.png')
A number of groups – including those from the commercial, governmental, and academic sectors have created knowledgebases to interpret cancer genomic mutations in the context of evidence for pathogenicity, relevant treatment options, or genomically-guided clinical trials (e.g., NCI MATCH and ASCO TAPUR basket trials, and many individual targeted therapy trials). However, in practice, clinicians and researchers are currently unable to effectively utilize the accumulated knowledge derived from such efforts. Integration of the available knowledge is infeasible because each institution (often redundantly) curates their own knowledgebase with limited adherence to any interoperability standards, contributing to a persistent data sharing and interpretation bottleneck. G2P is an aggregate public clinical cancer knowledge base for storing and searching connections between genomic biomarkers (“genotypes”) and patient diagnosis, prognosis, and response to treatment (“phenotypes”). Key uses of G2P include (a) searching by somatic variant to find drugs known to lead to response or resistance in tumors with the variant; (b) searching by drug to identify different mutations in which it can lead to response; (c) searching clinical trials to find those associated with particular biomarkers or drugs. G2P combines biomarker-phenotype associations from 9 trusted and curated knowledge bases, including:
- Jackson Lab Clinical Knowledge Base
- Washington University CIViC
- Precision Oncology Knowledge Base
- Cancer Genome Interpreter Cancer bioMarkers database
- GA4GH reference server
- Cornell pmkb
- MolecularMatch
- Clinical Trials: MolecularMatch and Jackson Lab
Key Accomplishments for this effort to date:
- Used internally at OHSU for tumor boards - SMMART Research at OCSSB This collaborative research project brings together a multi-disciplinary team from across OHSU to discover 'omic and architectural features that manifest with the development of resistance to cancer treatments.
- Selection as a GA4GH driver project. The real world experience derived from harmonizing disparate semi structured data informs the efforts of the Variant Interpretation for Cancer Consortium (VICC). Considerable progress has been made on these specific aims:
- Harmonize global efforts for clinical interpretation of cancer variants and precision medicine clinical trial curation.
- Standardize data model for variant interpretation, the association between Evidence, Features, Drugs, Diseases.
- Implement software systems to query across standardized knowledgebases.
- Implement web application for result exploration and report generation.
- In addition to SMMART research, we created a GENIE cohort comparison analysis in preparation for a paper submittal to Nature Genetics
Plans:
- Trials: Determine the minimal elements needed to define structured clinical trial eligibility criteria. We will develop a set of minimal elements needed for defining structured genomic and clinical eligibility criteria for cancer precision medicine clinical trials. We will also create algorithms to match patients to clinical trials based on genomic information.
- Evidence: Additional source knowledge bases (opentargets, TP53)
- Self service: In order to evangelize and increase dissemination we will create a service to annotate the user's MAF/VCF data.
- Cohorts: To prepare for additional cohort comparisons we are integrating with Biomedical Evidence Graph graph database. BMEG stores data in a large property graph and provides a query language, Ophion (implementing much of the gremlin spec), to perform traversals through this graph and collect data along the way. To obtain the highest quality analytical methods the BMEG team has partnered with Sage BioNetworks to organize several DREAM Challenges including: SMC-DNA, SMC-Het, SMC-RNA. Upon completion this will allow queries to access several other datasets (CCLE, TCGA, GTEX, CTDD, SANGER CANCER RX, PATHWAY COMMON, PFAM). This infrastructure will also be used in collaboration with CHOP's novel cohort Kids First
Challenges:
- Many groups are curating interpretations for which mutations are clinically relevant but their efforts are siloed, often redundant, and not interoperable. Therefore, there is a clear need to standardize and coordinate clinical-genomics curation efforts, and create a public community resource able to query the aggregated information. Specifically, we will coordinate variant interpretation and clinical trial curation activities according to the domain expertise of different groups/institutes. We will attempt to unify efforts and leverage domain-specific expertise to reduce redundant curation effort. This will require developing methods for coordinating curation efforts, quality assurance, and clinician engagement. Access is currently at the API and data layer. This creates a challenge for curation/QA/Clinician engagement. Remediations include development of a bespoke UI and integration with other front ends (civic).
Publications/Websites:
- http://cancervariants.org/
- https://github.com/ohsu-comp-bio/g2p-aggregator/
- https://g2p-ohsu.ddns.net
Dissemination:
- Multiple presentations on GA4GH calls
- Presentation at GA4GH Orlando conference
- Grant application
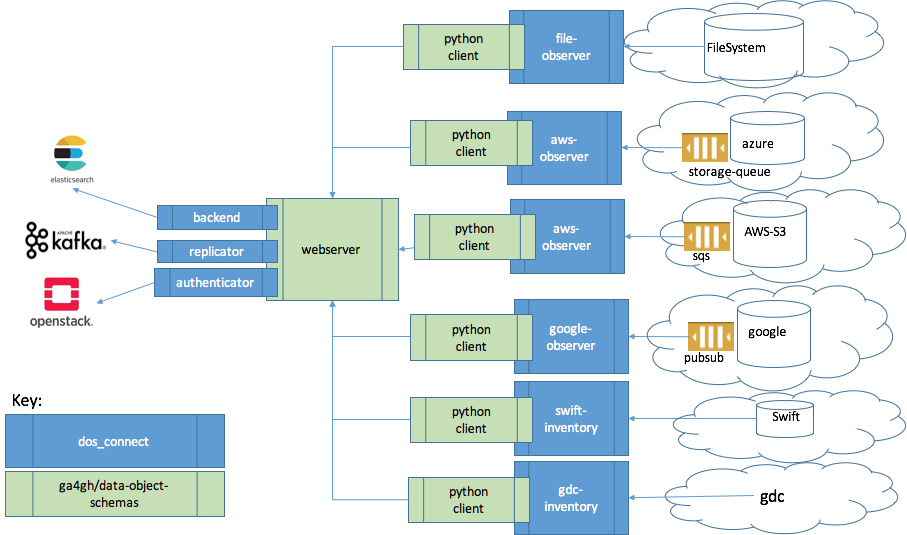
Through active collaboration with UCSC, University of Chicago and others, DOS provides a generic API, so workflow systems can access data in the same way regardless of project. One section of the API focuses on how to read and write data objects to cloud environments and how to join them together as data bundles (Data object management). Another focuses on the ability to find data objects across cloud environments and implementations of DOS (Data object queries). The latter is likely to be worked on in conjunction with the GA4GH Discovery Workstream.
The DOS Connect project, leverages the schemas and technologies developed by DOS provides two high level capabilities:
- observation: long lived services to observe the object store and populate a webserver with data-object-schema records. These observations catch add, moves and deletes to the object store.
- inventory: on demand commands to capture a snapshot of the object store using data-object-schema records.
Accomplishments:
- Established GA4GH schemas through active collaboration with UCSC
- Practical real-world implementation for multiple data sources including Swift, S3, Google Storage, Azure, File System, API (GDC)
Dissemination:
- Multiple presentations on GA4GH calls
- Presentation for CHOP Kids First
- Demonstration using OHSU's SMMART research datasets
Publications/Websites:
- https://github.com/ohsu-comp-bio/dos_connect
- https://github.com/ga4gh/data-object-service-schemas
Plans:
-
Create specific implementations for OICR/ICGC/HCA and Kids first and integrate back to those projects via pull requests to their repositories.
-
Explore and demonstrate how workflows FUNNEL/TES, Cromwell and others can use DOS' harmonized data to make decisions about what platform/region should be used to house data or excecute workflows.
-
Enabling specific 'success stories', for example DREAM SMC RNA-seq challenge as first multi-cloud deployment challenge.
Challenges:
- Scale: DOS addresses multi cloud projects, including those that span private clouds and site specific file systems. Many projects have addressed this in project specific "roll your own" alternatives. Remediations include evangelization, setting up a 'sandbox' where users can add their own object store buckets and plans for direct contributions back to key projects.