-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy pathREADME.Rmd
104 lines (74 loc) · 4.94 KB
/
README.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
---
output:
md_document:
variant: markdown_github
---
<!-- README.md is generated from README.Rmd. Please edit that file -->
```{r, echo = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "README-"
)
```
[](https://natverse.github.io)
[](https://travis-ci.org/natverse/fishatlas)
[](https://codecov.io/gh/natverse/fishatlas?branch=master)
[](http://jefferislab.github.io/fishatlas/reference/)
<img src="man/figures/logo.svg" align="right" height="139" />
# Fish Brain Atlas
The goal of *fishatlas* is to provide R client utilities for interacting with the [Fish Brain Atlas Project](https://www.neuro.mpg.de/baier/connectome), which has [successfully](https://www.cell.com/neuron/pdfExtended/S0896-6273(19)30391-5) acquired and registered almost 2,000 neurons from the larval zebrafish into a standard, annotated template space. Using this R package in concert with the [natverse](https://github.com/natverse/natverse) ecosystem of neuroanatomy tools is highly recommended. The [project](https://www.neuro.mpg.de/baier/connectome) was conducted by the group of [Herwig Baier](https://scholar.google.de/citations?user=e80hnfEAAAAJ&hl=en) at the [MPI](https://www.mpg.de/en), primarily by [Michael Kunst](https://scholar.google.co.uk/citations?user=yMyxCfQAAAAJ&hl=en). The website was created by [Nawwar Mokayes](https://www.linkedin.com/in/nouwarmokayes/?originalSubdomain=de). The larval brain data has all been collected from fish at 6 days post-fertilisation.
## Installation
Firstly, you will need R, R Studio and X Quartz as well as nat and its dependencies. For detailed installation instructions for all this, see [here](https://jefferis.github.io/nat/articles/Installation.html). It should not take too long at all. Then:
```{r install, eval = FALSE}
# install
if (!require("devtools")) install.packages("devtools")
devtools::install_github("natverse/fishatlas")
# use
library(fishatlas)
```
Done!
## Key Functions
Now we can have a look at what is available, here are some of the key functions. Their help details examples of their use. You can summon the help in RStudio using `?` followed by the function name.
```{r help, eval = FALSE}
# And how can I download and read neurons?
?fishatlas_read_saved_neurons()
# Get a 3D neuropil-subdivided brain model
?fishatlas_read_brain # Get 3D neuropil-subdivided brain models for those brainspaces
```
## Example
Let's also have a look at an example pulling neurons and brain meshes from [fishatlas.neuro.mpg.de](https://fishatlas.neuro.mpg.de). Excitingly, all the data is in a single standard template space!
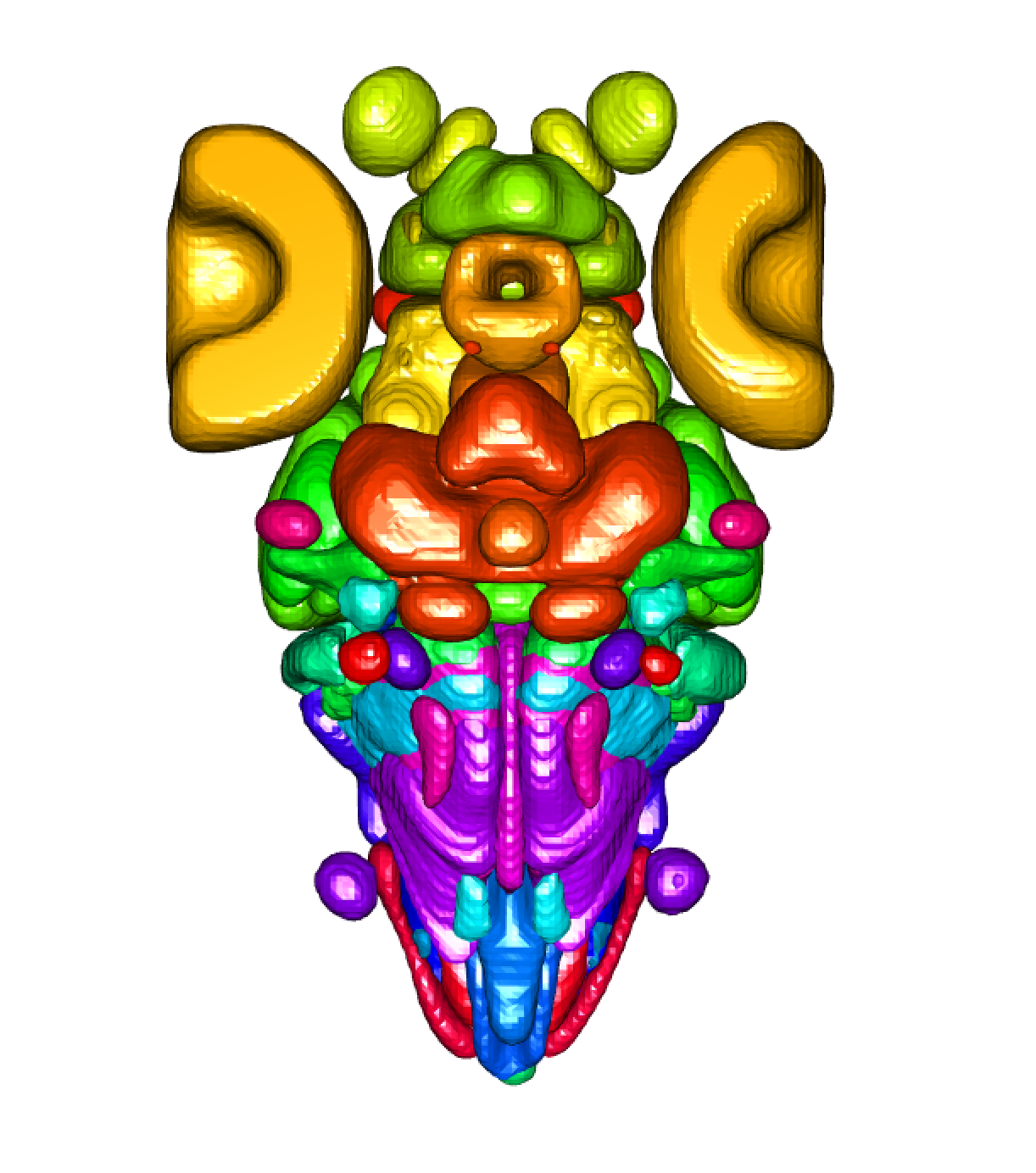
```{r insectbraindb.example, eval = FALSE}
## Lets have a look at these neuropils!
zfbrain = fishatlas_read_brain(type = "brain_regions")
plot3d(zfbrain)
## Maybe we just want to plot for forebrain
clear3d()
plot3d(subset(zfbrain, "Forebrain"), alpha = 0.5, col = "red")
## Maybe within the forebrain, we are interested in plotting the habenula
plot3d(subset(zfbrain, "Habenula"), alpha = 0.3, col = "red")
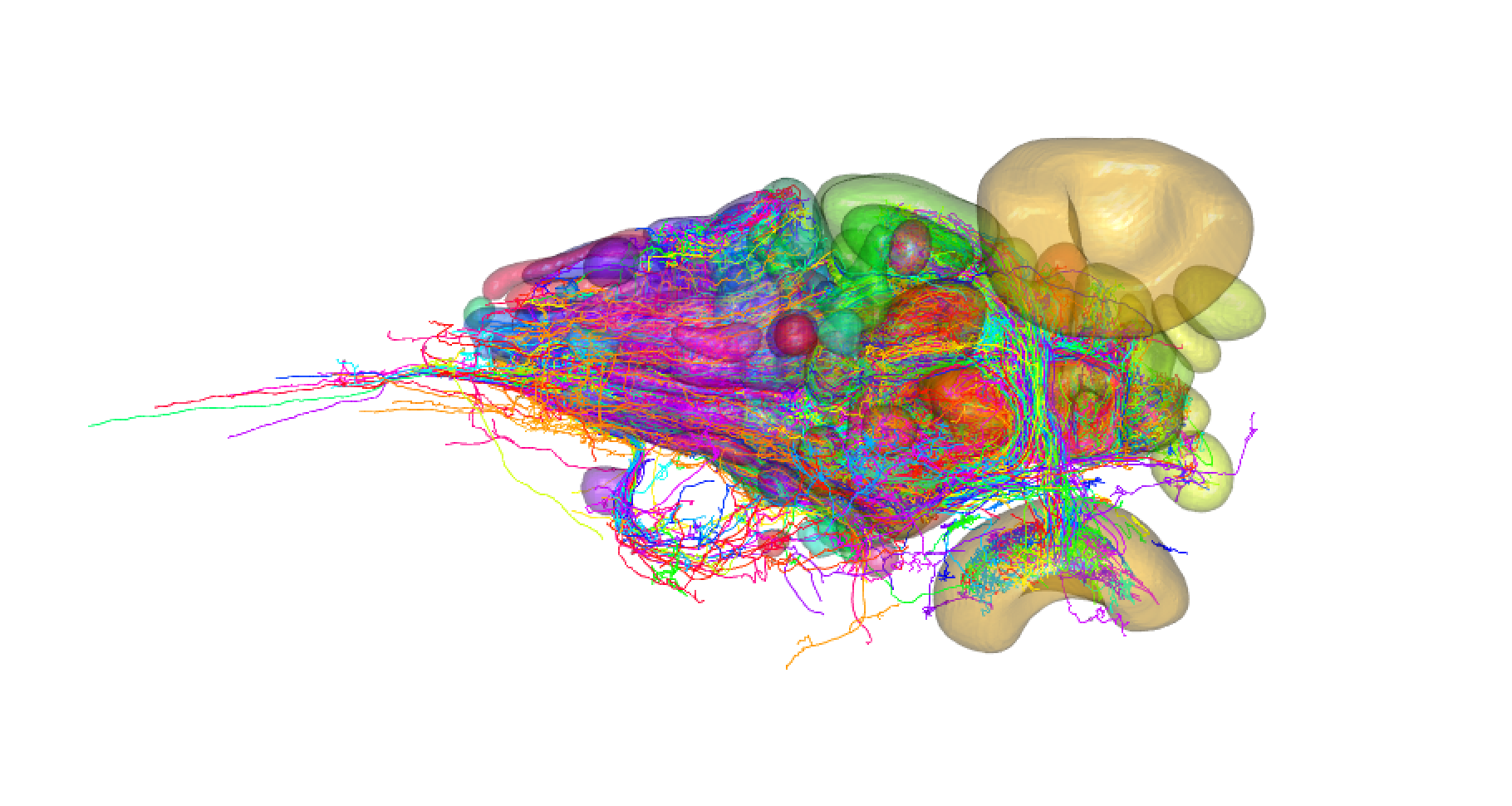
## First we need to download all of the neurons
### We should only ever have to do this once!
fishatlas_download_neurons()
## Let's get all that sweet neuron data!
zfishn = fishatlas_read_saved_neurons(side = "Original")
plot3d(zfishn,soma = TRUE, lwd = 2)
## Hmm, but it would be better to have them all on the same side
clear3d()
zfishr = fishatlas_read_saved_neurons(side = "Right")
plot3d(zfishr,soma = TRUE, lwd = 2, col = "red")
## Great! How does that compare with neurons all on the left?
### Let's just look at the first 10 mirored.
zfishl = fishatlas_mirror_saved_neurons(fishatlas_neurons[1:10], side = "Left")
plot3d(zfishl,soma = TRUE, lwd = 2, col = "cyan")
```
## Acknowledging the data and tools
Any work that uses data from this package should cite
**Kunst, Michael, Eva Laurell, Nouwar Mokayes, Anna Kramer, Fumi Kubo, António M. Fernandes, Dominique Förster, Marco Dal Maschio, and Herwig Baier.** 2019. *A Cellular-Resolution Atlas of the Larval Zebrafish Brain.* Neuron, May. https://doi.org/10.1016/j.neuron.2019.04.034.
This package was created by Alexander Shakeel Bates, while in the group of [Dr. Gregory Jefferis](https://en.wikipedia.org/wiki/Gregory_Jefferis). You can cite this package as:
```{r citation, eval = FALSE}
citation(package = "fishatlas")
```
**Bates AS** (2019). *fishatlas: R client utilities for interacting with the MPI Fish Brain Atlas project.* **R package** version 0.1.0. https://github.com/natverse/fishatlas